Biologists need computational tools that are easily scalable, customizable, and modular. I am moved by the concept of creating and accumulating all of the data gathering, formatting, and analysis modules that a particular lab might need such that even less computer-savvy researchers can easily customize their own workflows to do exactly what they want. Therefore, one of my main research interests is the development of robust, easy-to-use, open-source bioinformatic software.
I have several open-source coding projects of various sizes hosted on GitHub. Most are related to bioinformatics, while others are just fun side projects.
-
Maloof Lab CMS
Django django CMS Biodalliance Shiny Bootstrap 3
The Maloof Lab CMS comprises web apps for managing lab members, publications, genome browsers, Shiny apps, and other content relevant to a scientific lab.
-
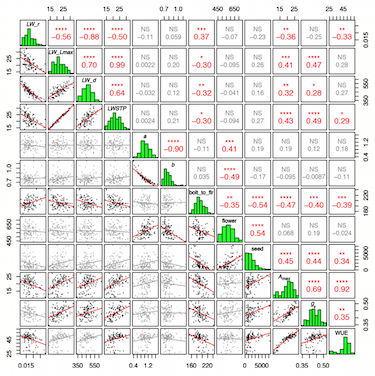
Modeling development and quantitative trait mapping reveal independent genetic modules for leaf size and shape.. New Phytologist (2015)
Baker RL, Leong WF, Brock MT, Markelz RJ, Covington MF, Devisetty UK, Edwards CE, Maloof J, Welch S, Weinig C
Improved predictions of fitness and yield may be obtained by characterizing the genetic controls and environmental dependencies of organismal ontogeny. Elucidating the shape of growth curves may reveal novel genetic controls that single-time-point (STP) analyses do not because, in theory, infinite numbers of growth curves can result in the same final measurement. We measured leaf lengths and widths in Brassica rapa recombinant inbred lines (RILs) throughout ontogeny. We modeled leaf growth and allometry as function valued traits (FVT), and examined genetic correlations between these traits and aspects of phenology, physiology, circadian rhythms and fitness. We used RNA-seq to construct a SNP linkage map and mapped trait quantitative trait loci (QTL). We found genetic trade-offs between leaf size and growth rate FVT and uncovered differences in genotypic and QTL correlations involving FVT vs STPs. We identified leaf shape (allometry) as a genetic module independent of length and width and identified selection on FVT parameters of development. Leaf shape is associated with venation features that affect desiccation resistance. The genetic independence of leaf shape from other leaf traits may therefore enable crop optimization in leaf shape without negative effects on traits such as size, growth rate, duration or gas exchange.
-
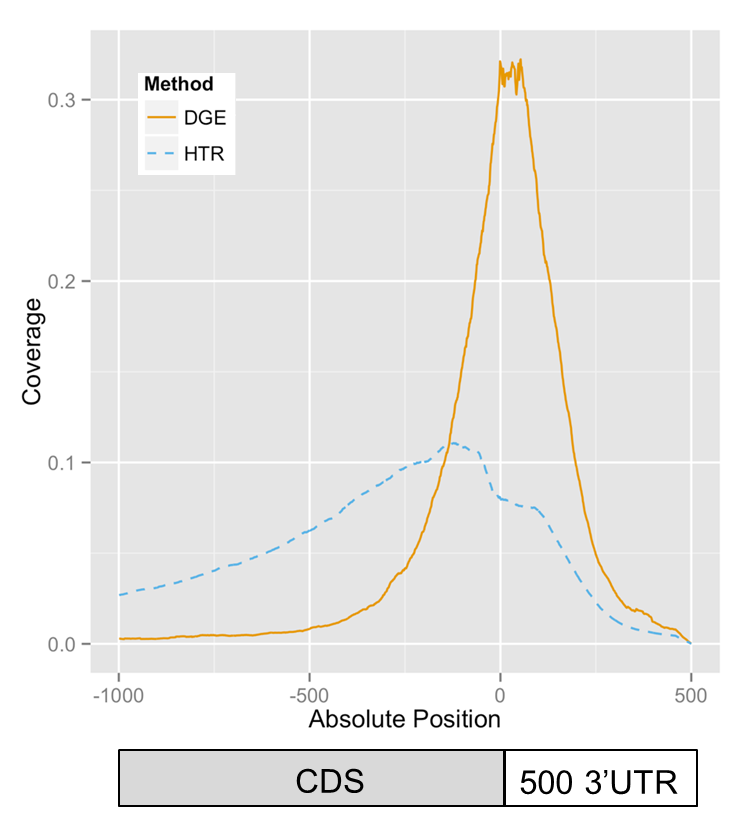
BrAD-seq: Breath Adapter Directional sequencing: a streamlined, ultra-simple and fast library preparation protocol for strand specific mRNA library construction. Frontiers in Plant Science (2015)
Townsley BT, Covington MF, Ichihashi Y, Zumstein Y, and Sinha NR
Next Generation Sequencing (NGS) is driving rapid advancement in biological understanding and RNA-sequencing (RNA-seq) has become an indispensable tool for biology and medicine. There is a growing need for access to these technologies although preparation of NGS libraries remains a bottleneck to wider adoption. Here we report a novel method for the production of strand specific RNA-seq libraries utilizing inherent properties of double-stranded cDNA to capture and incorporate a sequencing adapter. Breath Adapter Directional sequencing (BrAD-seq) reduces sample handling and requires far fewer enzymatic steps than most available methods to produce high quality strand-specific RNA-seq libraries. The method we present is optimized for 3-prime Digital Gene Expression (DGE) libraries and can easily extend to full transcript coverage shotgun (SHO) type strand-specific libraries and is modularized to accommodate a diversity of RNA and DNA input materials. BrAD-seq offers a highly streamlined and inexpensive option for RNA-seq libraries.
-
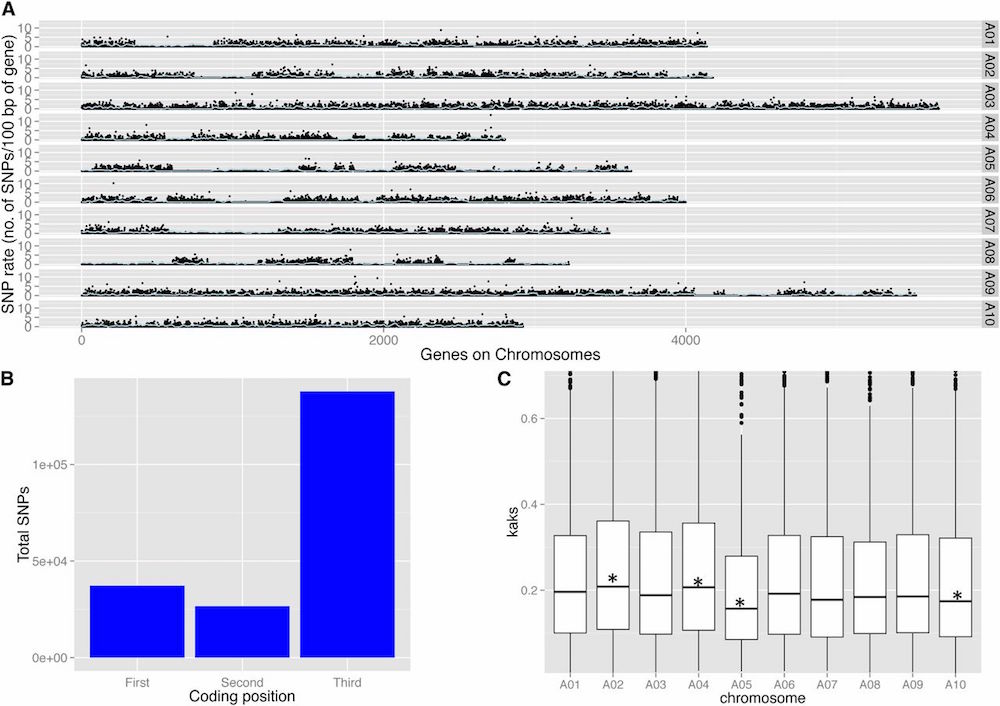
Polymorphism Identification and Improved Genome Annotation of Brassica rapa Through Deep RNA Sequencing. G3•Genes|Genomes|Genetics (2014)
Devisetty UK, Covington MF, Tat AV, Lekkala S, Maloof JN
The mapping and functional analysis of quantitative traits in Brassica rapa can be greatly improved with the availability of physically positioned, gene-based genetic markers and accurate genome annotation. In this study, deep transcriptome RNA sequencing (RNA-Seq) of Brassica rapa was undertaken with two objectives: SNP detection and improved transcriptome annotation. We performed SNP detection on two varieties that are parents of a mapping population to aid in development of a marker system for this population and subsequent development of high-resolution genetic map. An improved Brassica rapa transcriptome was constructed to detect novel transcripts and to improve the current genome annotation. This is useful for accurate mRNA abundance and detection of expression QTL (eQTLs) in mapping populations. Deep RNA-Seq of two Brassica rapa genotypes-R500 (var. trilocularis, Yellow Sarson) and IMB211 (a rapid cycling variety)-using eight different tissues (root, internode, leaf, petiole, apical meristem, floral meristem, silique, and seedling) grown across three different environments (growth chamber, greenhouse and field) and under two different treatments (simulated sun and simulated shade) generated 2.3 billion high-quality Illumina reads. A total of 330,995 SNPs were identified in transcribed regions between the two genotypes with an average frequency of one SNP in every 200 bases. The deep RNA-Seq reassembled Brassica rapa transcriptome identified 44,239 protein-coding genes. Compared with current gene models of B. rapa, we detected 3537 novel transcripts, 23,754 gene models had structural modifications, and 3655 annotated proteins changed. Gaps in the current genome assembly of B. rapa are highlighted by our identification of 780 unmapped transcripts. All the SNPs, annotations, and predicted transcripts can be viewed at http://phytonetworks.ucdavis.edu/.
-
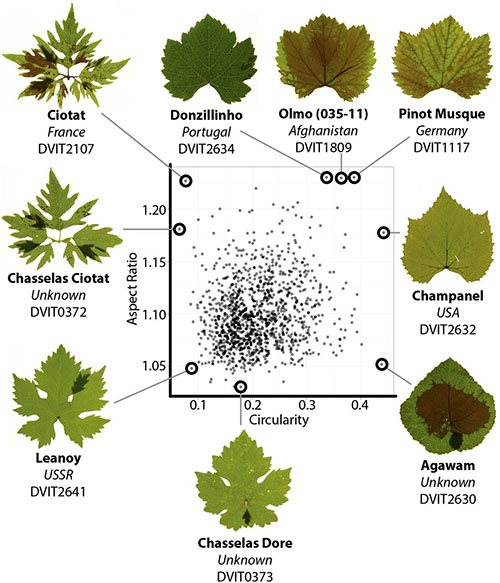
A modern ampelography: a genetic basis for leaf shape and venation patterning in grape. Plant Physiology (2014)
Chitwood DH, Ranjan A, Martinez CC, Headland LR, Thiem T, Kumar R, Covington MF, Hatcher T, Naylor DT, Zimmerman S, Downs N, Raymundo N, Buckler ES, Maloof JN, Aradhya M, Prins B, Li L, Myles S, Sinha NR
Terroir, the unique interaction between genotype, environment, and culture, is highly refined in domesticated grape (Vitis vinifera). Toward cultivating terroir, the science of ampelography tried to distinguish thousands of grape cultivars without the aid of genetics. This led to sophisticated phenotypic analyses of natural variation in grape leaves, which within a palmate-lobed framework exhibit diverse patterns of blade outgrowth, hirsuteness, and venation patterning. Here, we provide a morphometric analysis of more than 1,200 grape accessions. Elliptical Fourier descriptors provide a global analysis of leaf outlines and lobe positioning, while a Procrustes analysis quantitatively describes venation patterning. Correlation with previous ampelography suggests an important genetic component, which we confirm with estimates of heritability. We further use RNA-Seq of mutant varieties and perform a genome-wide association study to explore the genetic basis of leaf shape. Meta-analysis reveals a relationship between leaf morphology and hirsuteness, traits known to correlate with climate in the fossil record and extant species. Together, our data demonstrate a genetic basis for the intricate diversity present in grape leaves. We discuss the possibility of using grape leaves as a breeding target to preserve terroir in the face of anticipated climate change, a major problem facing viticulture.
-
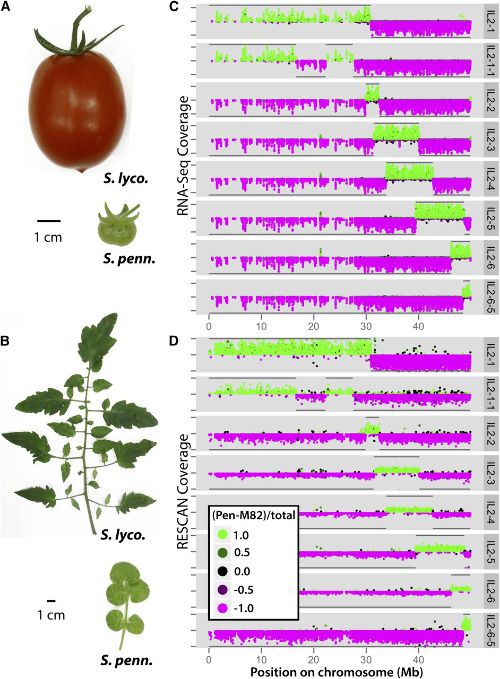
A Quantitative Genetic Basis for Leaf Morphology in a Set of Precisely Defined Tomato Introgression Lines. Plant Cell (2013)
Chitwood DH, Kumar R, Headland LR, Ranjan A, Covington MF, Ichihashi Y, Fulop D, Jiménez-Gómez JM, Peng J, Maloof JN, Sinha NR
PubMed PDF In Brief by J. Lockhart
Introgression lines (ILs), in which genetic material from wild tomato species is introgressed into a domesticated background, have been used extensively in tomato (Solanum lycopersicum) improvement. Here, we genotype an IL population derived from the wild desert tomato Solanum pennellii at ultrahigh density, providing the exact gene content harbored by each line. To take advantage of this information, we determine IL phenotypes for a suite of vegetative traits, ranging from leaf complexity, shape, and size to cellular traits, such as stomatal density and epidermal cell phenotypes. Elliptical Fourier descriptors on leaflet outlines provide a global analysis of highly heritable, intricate aspects of leaf morphology. We also demonstrate constraints between leaflet size and leaf complexity, pavement cell size, and stomatal density and show independent segregation of traits previously assumed to be genetically coregulated. Meta-analysis of previously measured traits in the ILs shows an unexpected relationship between leaf morphology and fruit sugar levels, which RNA-Seq data suggest may be attributable to genetically coregulated changes in fruit morphology or the impact of leaf shape on photosynthesis. Together, our results both improve upon the utility of an important genetic resource and attest to a complex, genetic basis for differences in leaf morphology between natural populations.
-
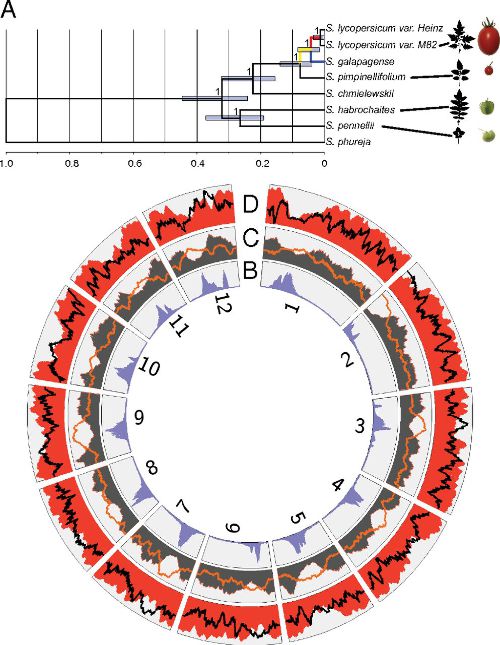
Comparative transcriptomics reveals patterns of selection in domesticated and wild tomato. Proc Natl Acad Sci USA (2013)
Koenig D, Jiménez-Gómez JM, Kimura S, Fulop D, Chitwood DH, Headland LR, Kumar R, Covington MF, Devisetty UK, Tat AV, Tohge T, Bolger A, Schneeberger K, Ossowski S, Lanz C, Xiong G, Taylor-Teeples M, Brady SM, Pauly M, Weigel D, Usadel B, Fernie AR, Peng J, Sinha NR, Maloof JN
Although applied over extremely short timescales, artificial selection has dramatically altered the form, physiology, and life history of cultivated plants. We have used RNAseq to define both gene sequence and expression divergence between cultivated tomato and five related wild species. Based on sequence differences, we detect footprints of positive selection in over 50 genes. We also document thousands of shifts in gene-expression level, many of which resulted from changes in selection pressure. These rapidly evolving genes are commonly associated with environmental response and stress tolerance. The importance of environmental inputs during evolution of gene expression is further highlighted by large-scale alteration of the light response coexpression network between wild and cultivated accessions. Human manipulation of the genome has heavily impacted the tomato transcriptome through directed admixture and by indirectly favoring nonsynonymous over synonymous substitutions. Taken together, our results shed light on the pervasive effects artificial and natural selection have had on the transcriptomes of tomato and its wild relatives.
-

Circadian control of jasmonates and salicylates: The clock role in plant defense. Plant Signal Behav. (2013)
Goodspeed D, Chehab EW, Covington MF, Braam J
Plants have evolved robust mechanisms to perceive and respond to diverse environmental stimuli. The plant phytohormones jasmonates and salicylates play key roles in activating biotic stress response pathways. Recent findings demonstrate that basal levels of both jasmonates and salicylates in Arabidopsis are under the control of the circadian clock and that clock-controlled jasmonate accumulation may underlie clock- and jasmonate-dependent enhanced resistance of Arabidopsis to Trichoplusia ni (cabbage looper), a generalist herbivore. Here we summarize these findings and provide further evidence that a functional plant circadian clock is required for optimal herbivore defense in Arabidopsis. When given a choice to feed on wild-type plants or arrhythmic transgenics, T. ni prefer plants lacking robust circadian rhythms. Altogether these data provide strong evidence for circadian clock enabling anticipation of herbivore attack and thus contributing to overall plant fitness.
-
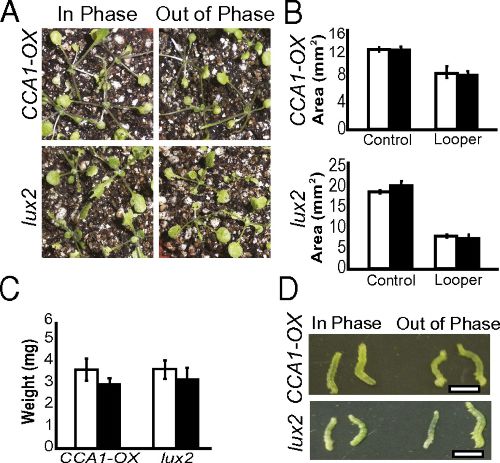
Arabidopsis synchronizes jasmonate-mediated defense with insect circadian behavior. Proc Natl Acad Sci USA (2012)
Goodspeed D, Chehab EW, Min-Venditti A, Braam J, Covington MF
PubMed PDF PDF + SI Commentary by G. Jander 2012 Cozzarelli Prize Recipient
Diverse life forms have evolved internal clocks enabling them to monitor time and thereby anticipate the daily environmental changes caused by Earth's rotation. The plant circadian clock regulates expression of about one-third of the Arabidopsis genome, yet the physiological relevance of this regulation is not fully understood. Here we show that the circadian clock, acting with hormone signals, provides selective advantage to plants through anticipation of and enhanced defense against herbivory. We found that cabbage loopers (Trichoplusia ni) display rhythmic feeding behavior that is sustained under constant conditions, and plants entrained in light/dark cycles coincident with the entrainment of the T. ni suffer only moderate tissue loss due to herbivory. In contrast, plants entrained out-of-phase relative to the insects are significantly more susceptible to attack. The in-phase entrainment advantage is lost in plants with arrhythmic clocks or deficient in jasmonate hormone; thus, both the circadian clock and jasmonates are required. Circadian jasmonate accumulation occurs in a phase pattern consistent with preparation for the onset of peak circadian insect feeding behavior, providing evidence for the underlying mechanism of clock-enhanced herbivory resistance. Furthermore, we find that salicylate, a hormone involved in biotrophic defense that often acts antagonistically to jasmonates, accumulates in opposite phase to jasmonates. Our results demonstrate that the plant circadian clock provides a strong physiological advantage by performing a critical role in Arabidopsis defense.
-
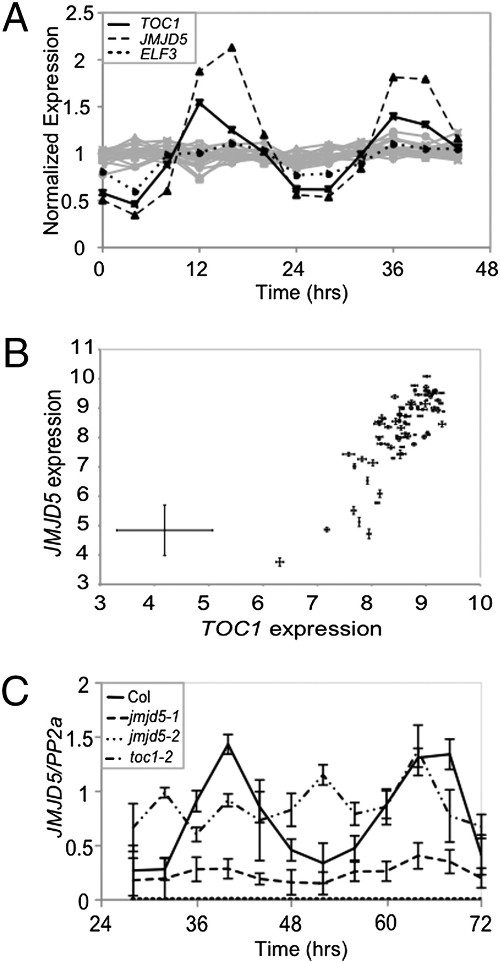
Jumonji domain protein JMJD5 functions in both the plant and human circadian systems. Proc Natl Acad Sci USA (2010)
Jones MA, Covington MF, DiTacchio L, Vollmers C, Panda S, Harmer SL
Circadian clocks are near-ubiquitous molecular oscillators that coordinate biochemical, physiological, and behavioral processes with environmental cues, such as dawn and dusk. Circadian timing mechanisms are thought to have arisen multiple times throughout the evolution of eukaryotes but share a similar overall structure consisting of interlocking transcriptional and posttranslational feedback loops. Recent work in both plants and animals has also linked modification of histones to circadian clock function. Now, using data from published microarray experiments, we have identified a histone demethylase, jumonji domain containing 5 (JMJD5), as a previously undescribed participant in both the human and Arabidopsis circadian systems. Arabidopsis JMJD5 is coregulated with evening-phased clock components and positively affects expression of clock genes expressed at dawn. We found that both Arabidopsis jmjd5 mutant seedlings and mammalian cell cultures deficient for the human ortholog of this gene have similar fast-running circadian oscillations compared with WT. Remarkably, both the Arabidopsis and human JMJD5 orthologs retain sufficient commonality to rescue the circadian phenotype of the reciprocal system. Thus, JMJD5 plays an interchangeable role in the timing mechanisms of plants and animals despite their highly divergent evolutionary paths.
-
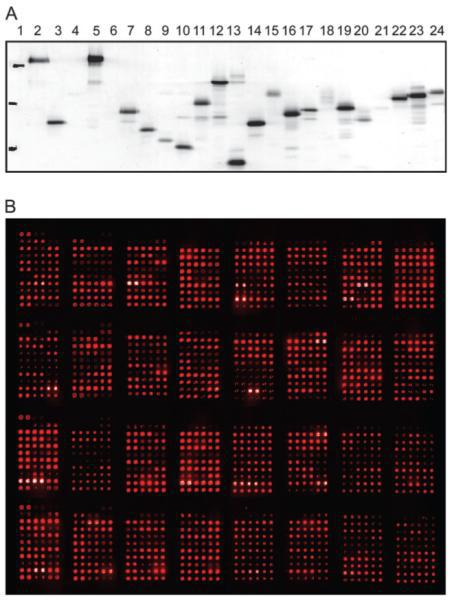
The development of protein microarrays and their applications in DNA-protein and protein-protein interaction analyses of Arabidopsis transcription factors. Mol Plant (2008)
Gong W, He K, Covington MF, Dinesh-Kumar SP, Snyder M, Harmer SL, Zhu YX, Deng XW.
We used our collection of Arabidopsis transcription factor (TF) ORFeome clones to construct protein microarrays containing as many as 802 TF proteins. These protein microarrays were used for both protein-DNA and protein-protein interaction analyses. For protein-DNA interaction studies, we examined AP2/ERF family TFs and their cognate cis-elements. By careful comparison of the DNA-binding specificity of 13 TFs on the protein microarray with previous non-microarray data, we showed that protein microarrays provide an efficient and high throughput tool for genome-wide analysis of TF-DNA interactions. This microarray protein-DNA interaction analysis allowed us to derive a comprehensive view of DNA-binding profiles of AP2/ERF family proteins in Arabidopsis. It also revealed four TFs that bound the EE (evening element) and had the expected phased gene expression under clock-regulation, thus providing a basis for further functional analysis of their roles in clock regulation of gene expression. We also developed procedures for detecting protein interactions using this TF protein microarray and discovered four novel partners that interact with HY5, which can be validated by yeast two-hybrid assays. Thus, plant TF protein microarrays offer an attractive high-throughput alternative to traditional techniques for TF functional characterization on a global scale.
-
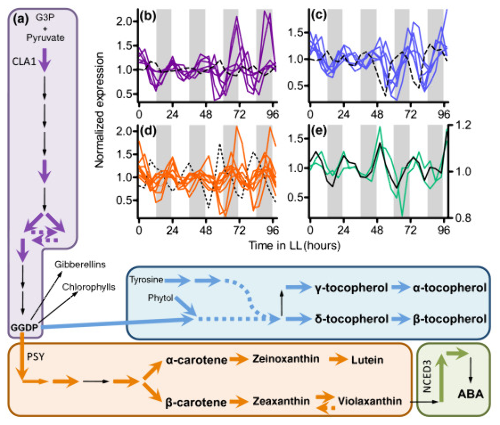
Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. (2008)
Covington MF, Maloof JN, Straume M, Kay SA, Harmer SL
BACKGROUND: As nonmotile organisms, plants must rapidly adapt to ever-changing environmental conditions, including those caused by daily light/dark cycles. One important mechanism for anticipating and preparing for such predictable changes is the circadian clock. Nearly all organisms have circadian oscillators that, when they are in phase with the Earth's rotation, provide a competitive advantage. In order to understand how circadian clocks benefit plants, it is necessary to identify the pathways and processes that are clock controlled.
RESULTS: We have integrated information from multiple circadian microarray experiments performed on Arabidopsis thaliana in order to better estimate the fraction of the plant transcriptome that is circadian regulated. Analyzing the promoters of clock-controlled genes, we identified circadian clock regulatory elements correlated with phase-specific transcript accumulation. We have also identified several physiological pathways enriched for clock-regulated changes in transcript abundance, suggesting they may be modulated by the circadian clock.
CONCLUSION: Our analysis suggests that transcript abundance of roughly one-third of expressed A. thaliana genes is circadian regulated. We found four promoter elements, enriched in the promoters of genes with four discrete phases, which may contribute to the time-of-day specific changes in the transcript abundance of these genes. Clock-regulated genes are over-represented among all of the classical plant hormone and multiple stress response pathways, suggesting that all of these pathways are influenced by the circadian clock. Further exploration of the links between the clock and these pathways will lead to a better understanding of how the circadian clock affects plant growth and leads to improved fitness.
-
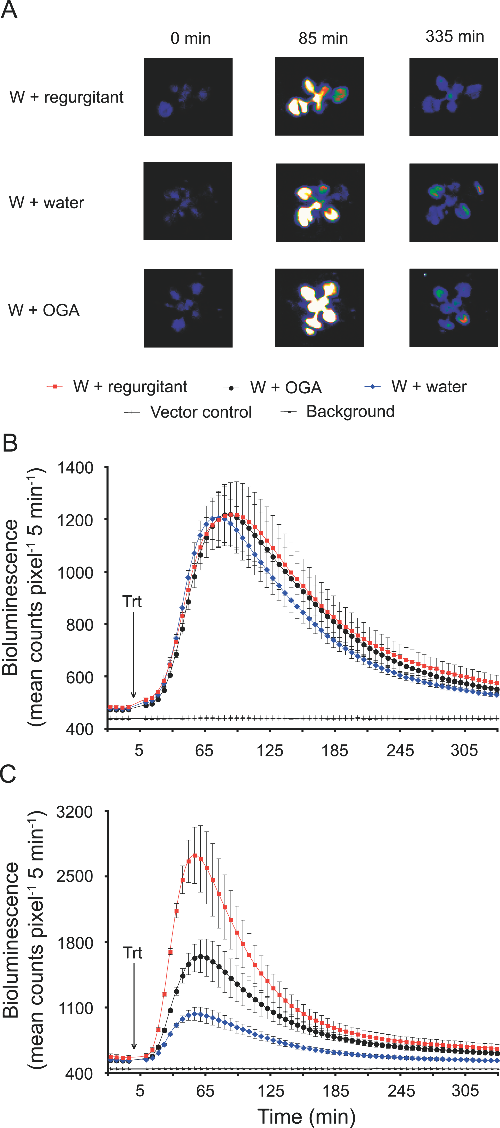
Mechanical stress induces biotic and abiotic stress responses via a novel cis-element. PLoS Genet. (2007)
Walley JW, Coughlan S, Hudson ME, Covington MF, Kaspi R, Banu G, Harmer SL, Dehesh K
Plants are continuously exposed to a myriad of abiotic and biotic stresses. However, the molecular mechanisms by which these stress signals are perceived and transduced are poorly understood. To begin to identify primary stress signal transduction components, we have focused on genes that respond rapidly (within 5 min) to stress signals. Because it has been hypothesized that detection of physical stress is a mechanism common to mounting a response against a broad range of environmental stresses, we have utilized mechanical wounding as the stress stimulus and performed whole genome microarray analysis of Arabidopsis thaliana leaf tissue. This led to the identification of a number of rapid wound responsive (RWR) genes. Comparison of RWR genes with published abiotic and biotic stress microarray datasets demonstrates a large overlap across a wide range of environmental stresses. Interestingly, RWR genes also exhibit a striking level and pattern of circadian regulation, with induced and repressed genes displaying antiphasic rhythms. Using bioinformatic analysis, we identified a novel motif overrepresented in the promoters of RWR genes, herein designated as the Rapid Stress Response Element (RSRE). We demonstrate in transgenic plants that multimerized RSREs are sufficient to confer a rapid response to both biotic and abiotic stresses in vivo, thereby establishing the functional involvement of this motif in primary transcriptional stress responses. Collectively, our data provide evidence for a novel cis-element that is distributed across the promoters of an array of diverse stress-responsive genes, poised to respond immediately and coordinately to stress signals. This structure suggests that plants may have a transcriptional network resembling the general stress signaling pathway in yeast and that the RSRE element may provide the key to this coordinate regulation.
-
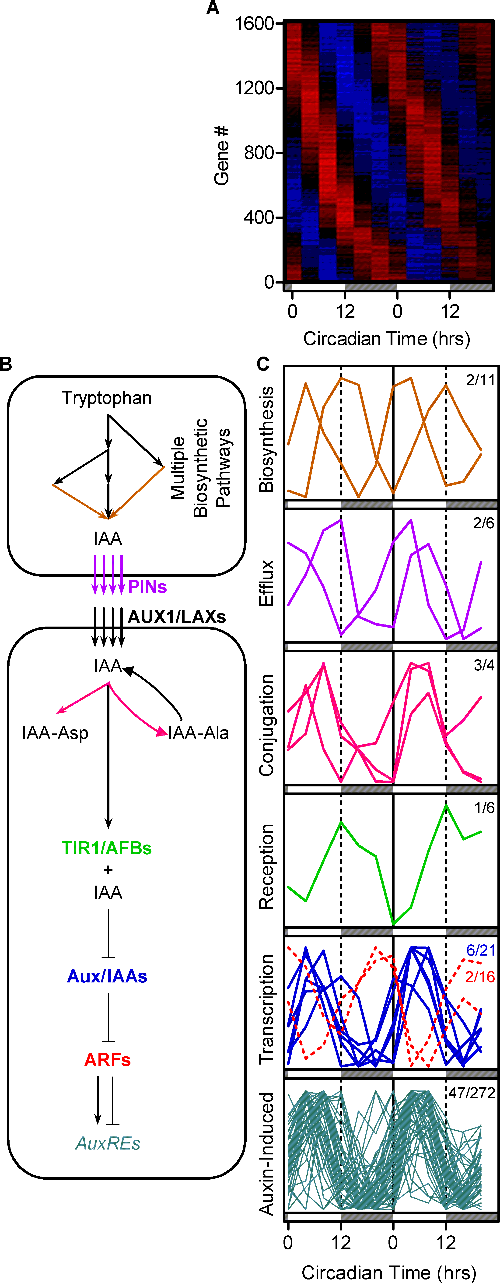
The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol. (2007)
Covington MF, Harmer SL
PubMed PDF Synopsis by M. Hoff
The circadian clock plays a pervasive role in the temporal regulation of plant physiology, environmental responsiveness, and development. In contrast, the phytohormone auxin plays a similarly far-reaching role in the spatial regulation of plant growth and development. Went and Thimann noted 70 years ago that plant sensitivity to auxin varied according to the time of day, an observation that they could not explain. Here we present work that explains this puzzle, demonstrating that the circadian clock regulates auxin signal transduction. Using genome-wide transcriptional profiling, we found many auxin-induced genes are under clock regulation. We verified that endogenous auxin signaling is clock regulated with a luciferase-based assay. Exogenous auxin has only modest effects on the plant clock, but the clock controls plant sensitivity to applied auxin. Notably, we found both transcriptional and growth responses to exogenous auxin are gated by the clock. Thus the circadian clock regulates some, and perhaps all, auxin responses. Consequently, many aspects of plant physiology not previously thought to be under circadian control may show time-of-day-specific sensitivity, with likely important consequences for plant growth and environmental responses.
-
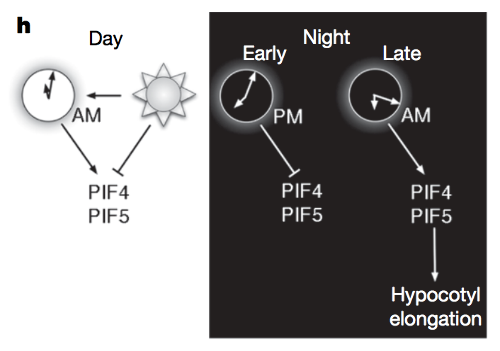
Rhythmic growth explained by coincidence between internal and external cues. Nature (2007)
Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL, Maloof JN
PubMed PDF News & Views by G. Breton and S. Kay
Most organisms use circadian oscillators to coordinate physiological and developmental processes such as growth with predictable daily environmental changes like sunrise and sunset. The importance of such coordination is highlighted by studies showing that circadian dysfunction causes reduced fitness in bacteria and plants, as well as sleep and psychological disorders in humans. Plant cell growth requires energy and water-factors that oscillate owing to diurnal environmental changes. Indeed, two important factors controlling stem growth are the internal circadian oscillator and external light levels. However, most circadian studies have been performed in constant conditions, precluding mechanistic study of interactions between the clock and diurnal variation in the environment. Studies of stem elongation in diurnal conditions have revealed complex growth patterns, but no mechanism has been described. Here we show that the growth phase of Arabidopsis seedlings in diurnal light conditions is shifted 8-12 h relative to plants in continuous light, and we describe a mechanism underlying this environmental response. We find that the clock regulates transcript levels of two basic helix-loop-helix genes, phytochrome-interacting factor 4 (PIF4) and PIF5, whereas light regulates their protein abundance. These genes function as positive growth regulators; the coincidence of high transcript levels (by the clock) and protein accumulation (in the dark) allows them to promote plant growth at the end of the night. Thus, these two genes integrate clock and light signalling, and their coordinated regulation explains the observed diurnal growth rhythms. This interaction may serve as a paradigm for understanding how endogenous and environmental signals cooperate to control other processes.
-
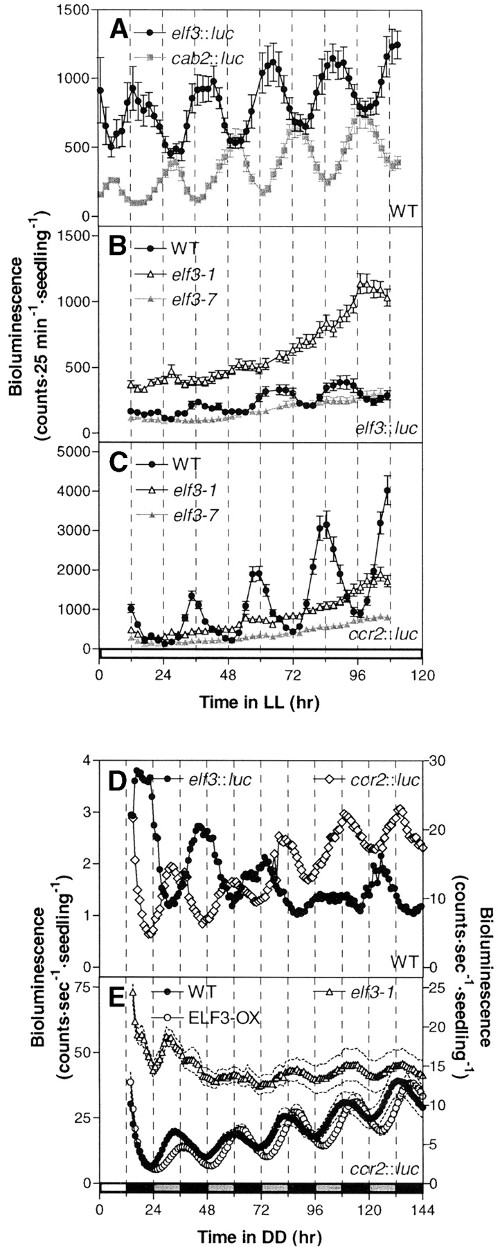
ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell (2001)
Covington MF, Panda S, Liu XL, Strayer CA, Wagner DR, Kay SA
The Arabidopsis early flowering 3 (elf3) mutation causes arrhythmic circadian output in continuous light, but there is some evidence of clock function in darkness. Here, we show conclusively that normal circadian function occurs with no alteration of period length in elf3 mutants in dark conditions and that the light-dependent arrhythmia observed in elf3 mutants is pleiotropic on multiple outputs normally expressed at different times of day. Plants overexpressing ELF3 have an increased period length in both constant blue and red light; furthermore, etiolated ELF3-overexpressing seedlings exhibit a decreased acute CAB2 response after a red light pulse, whereas the null mutant is hypersensitive to acute induction. This finding suggests that ELF3 negatively regulates light input to both the clock and its outputs. To determine whether ELF3's action is phase dependent, we examined clock resetting by using light pulses and constructed phase response curves. Absence of ELF3 activity causes a significant alteration of the phase response curve during the subjective night, and constitutive overexpression of ELF3 results in decreased sensitivity to the resetting stimulus, suggesting that ELF3 antagonizes light input to the clock during the night. The phase of ELF3 function correlates with its peak expression levels in the subjective night. ELF3 action, therefore, represents a mechanism by which the oscillator modulates light resetting.
-
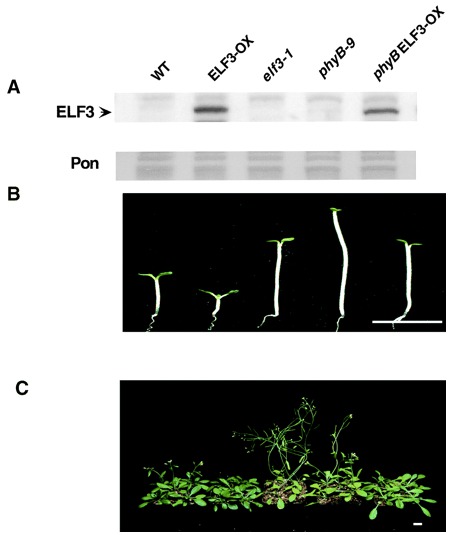
ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell (2001)
Liu XL, Covington MF, Fankhauser C, Chory J, Wagner DR
Many aspects of plant development are regulated by photoreceptor function and the circadian clock. Loss-of-function mutations in the Arabidopsis EARLY FLOWERING 3 (ELF3) and PHYTOCHROME B (PHYB) genes cause early flowering and influence the activity of circadian clock-regulated processes. We demonstrate here that the relative abundance of the ELF3 protein, which is a novel nucleus-localized protein, displays circadian regulation that follows the pattern of circadian accumulation of ELF3 transcript. Furthermore, the ELF3 protein interacts with PHYB in the yeast two-hybrid assay and in vitro. Genetic analyses show that ELF3 requires PHYB function in early morphogenesis but not for the regulation of flowering time. This suggests that ELF3 is a component of a PHYB signaling complex that controls early events in plant development but that ELF3 and PHYB control flowering via independent signal transduction pathways.
Mike Covington
Assistant Project Scientist
Maloof Lab
Section of Plant Biology
UC-Davis
mfcovington@gmail.com
GitHub/mfcovington
PerlMonks/frozenwithjoy StackExchange/mfcovington